Concept placement using BERT for biomedical ontology
we propose a method to automatically predict the presence of IS-A relationships between a new concept and pre-existing concepts based on the language representation model BERT.
The paper of this project is available here.
General idea of Transfer Learning with Google BERT
The idea of transfer learning is to use the knowledge learned from one task, and use it for another task. For example, we can take the a “pre-trained” CNN model trained using all the labeled general images (a large dataset) on ImageNet, and “fine-tune” it as a dog/cat classifier with our own training data (small dataset). Pre-training is normally expensive, because the model typically has a large number of parameters that need to be trained from scratch with a large size training data. Fine-tuning is normally cheap, because we will reuse the pre-trained model and exploit the “knowledge” it already learned, by training it with a small training dataset for our task-of-interest.
However, if we want, we can continue pre-training the model pre-trained by others. For example, the model we used here is BERT, which is pre-trained by Google using Wikipedia and BookCorpus text data, denoted as BERT_base. If we want to give the model a “medical flavor,” we can adopt the same training process Google did, but training BERT_base with medical domain text data. We can refer to the obtained model as BERT_base_medical. Note that here we are still in the scope of pre-training. We do not bind our model with any specific task. After we obtain the BERT_base_medical model, we can add a classifier/regression layer on top of it, say BERT_base_medical_classifier. Then we can fine-tune this new model with the training data for our task.

Concept placement using BERT
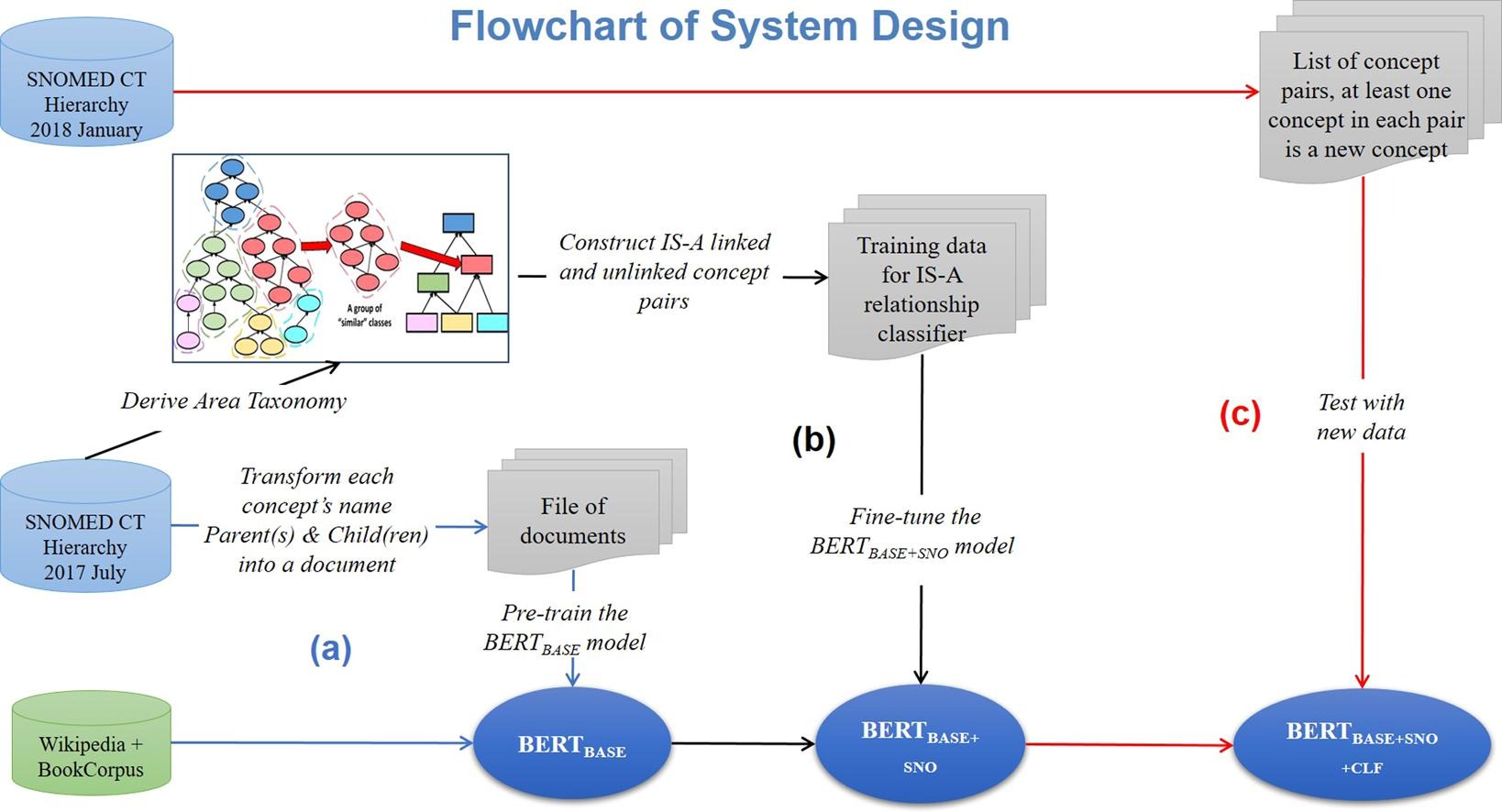
- We trained our model with the two largest hierarchies of the SNOMED CT 2017 July release and applied it to predicting the parents of new concepts added in the SNOMED CT 2018 January release.
- The results showed that our method achieved an average F1 score of 0.88, and the average Recall score improves slightly from 0.94 to 0.96 by using the ontology summarization technique.
In this project, we consider combining the two improvements by utilizing ontology summarization together with the BERT model and with an improved presentation of the training data to better utilize the “next sentence prediction” capability of BERT. We demonstrated that the language representation model BERT can be fine-tuned to predict IS-A relationships between new concepts and pre-existing concepts in SNOMED CT. This model can not only identify potential parents of a new concept, but also filter out irrelevant concepts, reducing the number of improper placement choices for a concept. We showed that the trained BERT model achieved an average F1 (F2) score of 0.87 (0.92) in testing with 8,574 concept pairs containing 2005 new concepts in the Clinical Finding hierarchy of SNOMED CT. The average F1 (F2) score in testing with 3,908 concept pairs containing 911 new concepts for the Procedure hierarchy was 0.821 (0.912). Furthermore, we employed the Area Taxonomy ontology summarization technique to refine the training data, which resulted in a higher Recall. Ontology curators can benefit from this high Recall, since it indicates that the trained model will propose a higher ratio of proper parents for a given concept. Therefore, the proposed method can save curators time and effort that would be needed to search for those parent candidates manually.

The source code of this project is available at https://github.com/hl395/Bert_Ontology